|
|
Welcome to The Visible Embryo, a comprehensive educational resource on human development from conception to birth.
The Visible Embryo provides visual references for changes in fetal development throughout pregnancy and can be navigated via fetal development or maternal changes.
The National Institutes of Child Health and Human Development awarded Phase I and Phase II Small Business Innovative Research Grants to develop The Visible Embryo. Initally designed to evaluate the internet as a teaching tool for first year medical students, The Visible Embryo is linked to over 600 educational institutions and is viewed by more than one million visitors each month.
Today, The Visible Embryo is linked to over 600 educational institutions and is viewed by more than 1 million visitors each month. The field of early embryology has grown to include the identification of the stem cell as not only critical to organogenesis in the embryo, but equally critical to organ function and repair in the adult human. The identification and understanding of genetic malfunction, inflammatory responses, and the progression in chronic disease, begins with a grounding in primary cellular and systemic functions manifested in the study of the early embryo.

The World Health Organization (WHO) has created a new Web site to help researchers, doctors and
patients obtain reliable information on high-quality clinical trials. Now you can go to one website and search all registers to identify clinical trial research underway around the world!

|
|
| Disclaimer: The Visible Embryo web site is provided for your general information only. The information contained on this site should not be treated as a substitute for medical, legal or other professional advice. Neither is The Visible Embryo responsible or liable for the contents of any websites of third parties which are listed on this site. |
|
|
 |
|
Content protected under a Creative Commons License. Commons License.
No dirivative works may be made or used for commercial purposes. |
|
|
| |
|
|

CLICK ON weeks 0 - 40 and follow along every 2 weeks of fetal development
|
|
|
|
|
Home | Pregnancy Timeline | News Alerts |News Archive Dec 1, 2014
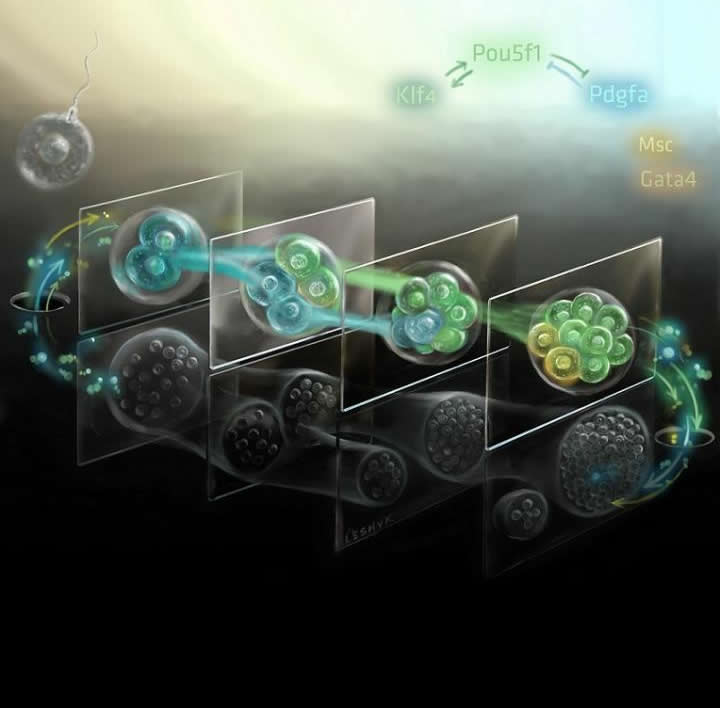
Bioengineering researchers used single-cell RNA-sequencing to measure each gene in a mouse genome at multiple stages of development to find which genes are being expressed at precise stages.
Image Credit: Art by Victor O. Leshyk courtesy of Sheng Zhong,
University of California San Diego Jacobs School of Engineering.
|
|
|
|
|
|
Two-cell embryos already "talk" about their future
Bioengineers have uncovered that mouse embryos immediately make decisions on their cell fate when only two to four cells in number. This discovery challenges the scientific consensus of when embryonic cells begin differentiating into cell types.
Research leader Sheng Zhong's research is in the field of systems, or network, biology and applies engineering principals to how biological systems work. For example, his team developed analytical methods to predict personal phenotypes — those observable characteristics we see that define a person's uniqueness ranging from eye and hair color to health and disposition — from that person's genome and epigenome.
Epigenome refers to all the chemical compounds regulating gene expression which vary from person to person. Predicting phenotypes by genome and epigenome is an emerging field in personalized medicine. Scientists believe an individual's epigenome could one day provide new ways to predict and treat genetic disorders.
The University of California, San Diego (UCSD) researchers used single-cell RNA sequencing to examine every gene in a mouse genome. Their work was published in the journal Genome Research. Additionally, the same group published an analysis of a single-cell's "time-course" registering the precise stages of embryonic development in the journal Proceedings of the National Academy of Sciences.
"Until recently, we didn't have the technology to look at cells this closely. Using single-cell RNA-sequencing, we are able to measure every gene in the mouse genome at multiple stages of development to find differences in gene expression at precise stages."
Sheng Zhong PhD, bioengineering professor, UCSD Jacobs School of Engineering, research leader.
These findings reveal cell activity that could provide insight into where normal development breaks down and early miscarriage and/or birth defects occur.
The scientists discovered that a handful of genes are clearly signaling to each other at the two and four-cell stages immediately following fertilization and before an embryo has implanted into the wall of the uterus. Among the identified genes are several belonging to the WNT signaling pathway, well-known for a role in cell-cell communications.
The prevailing view until now has been that mammalian embryos begin differentiating into cell types based upon their mass or volume of cells. According to co-authors Fernando Biase and Xiaoyi Cao, the first cell fate decision is still an open question. But, the first major objective of any embryo is still which cells will form the fetus, and which the placenta.
Genome Research
Abstract
It remains an open question when and how the first cell fate decision is made in mammals. Using deep single-cell RNA-seq of matched sister blastomeres, we report highly reproducible inter-blastomere differences among ten 2-cell and five 4-cell mouse embryos. Inter-blastomere gene expression differences dominated between-embryo differences and noises, and were sufficient to cluster sister blastomeres into distinct groups. Dozens of protein-coding genes exhibited reproducible bimodal expression in sister blastomeres (0 vs. 103-106 of FPKM), which cannot be explained by random fluctuations. The protein expression of one, out of four of these bimodal genes tested, Gadd45a, exhibited clear inter-blastomeric contrasts. We traced some of the bimodal mRNA expressions to embryonic genome activation, and others to blastomere-specific RNA depletion. Inter-blastomere differences created co-expression gene networks that were much stronger and larger than those that can be possibly created by random noises. The highly correlated gene pairs at the 4-cell stage overlapped with those showing the same directions of differential expression between inner cell mass (ICM) and trophectoderm (TE). These data substantiate the hypothesis of inter-blastomere differences in 2- and 4-cell mouse embryos, and associate these differences with ICM/TE differences.
Proceedings of the National Academy of Sciences
Significance
Clustering, in essence, was a tool for describing spatial characteristics in the objects of concern. However, from social to biological studies, researchers’ interests in temporal characteristics often rival their interests in spatial features. Previous efforts to incorporate time information into clustering primarily relied on coding the time information into similarity metrics, thus reducing the problem into the classical paradigm of spatial clustering. The limitation is that the clustering outcomes are often time invariant. Here, we initiate a class of statistical methods that simultaneously infer spatial and temporal groupings. Such methods explicitly model the time dependencies of clustering indices over time. Our method inferred three genes to be associated with the earliest cell fate decision, which was corroborated by experimental validations.
Abstract
Both spatial characteristics and temporal features are often the subjects of concern in physical, social, and biological studies. This work tackles the clustering problems for time course data in which the cluster number and clustering structure change with respect to time, dubbed time-variant clustering. We developed a hierarchical model that simultaneously clusters the objects at every time point and describes the relationships of the clusters between time points. The hidden layer of this model is a generalized form of branching processes. A reversible-jump Markov Chain Monte Carlo method was implemented for model inference, and a feature selection procedure was developed. We applied this method to explore an open question in preimplantation embryonic development. Our analyses using single-cell gene expression data suggested that the earliest cell fate decision could start at the 4-cell stage in mice, earlier than the commonly thought 8- to 16-cell stage. These results together with independent experimental data from single-cell RNA-seq provided support against a prevailing hypothesis in mammalian development.
The research was funded by the National Institutes of Health (DP2OD007417) and the March of Dimes Foundation. The publications are "Cell fate inclination within 2-cell and 4-cell mouse embryos revealed by single-cell RNA sequencing", Genome Research, November 2014 and "Time-variant clustering model for understanding cell fate decisions", PNAS, November 2014.
Return to top of page |
