|
|
|
Home | Pregnancy Timeline | News Alerts |News Archive Apr 27, 2015
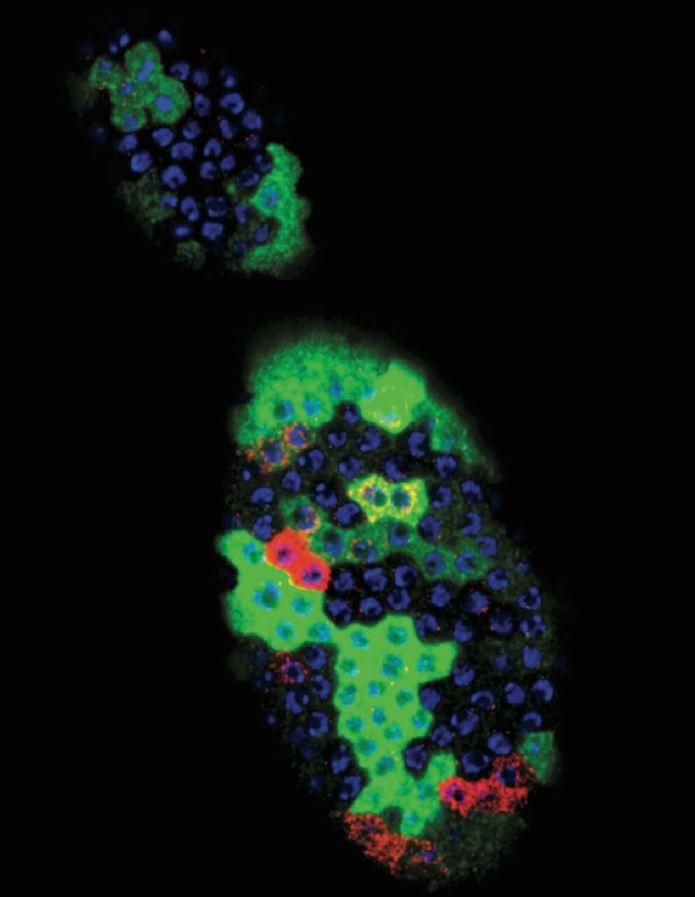
The ovary of the fruit fly with its' follicle progenitor cells in colors marking different cell stages —
(RED) specialized cells; (GREEN) gene expression; and (blue) nuclear DNA.
Image Credit: Ming-Chia Lee and Allan Spradling |
|
|
|
|
|
How young cells learn to communicate
Most developmental biologists have assumed that young cells, only recently born from stem cells and called "progenitors," are already competent at inter-communication with other cells - but they have a lot to learn.
New research from Allan Spradling of the Carnegie Institute and postdoctoral fellow Ming-Chia Lee, shows that infant progenitor cells go through a developmental process before advancing to become specific tissues. The existence of a naive stem cell state changes our understanding of how gene activity affects chromosome change.
The work is published in Genes and Development.
Molecules that package chromosomes to fit within the nucleus of a cell, control gene activity in what is known as the cell's "epigenetic state." To developmental biologists, changes in the epigenetic state ultimately explain how a cell is altered as it matures into a specific tissue.
"Acquired epigenetic changes in a developing cell are reminiscent of how learning changes the brain during childhood. Just as it remains difficult to map exactly what happens in a child's brain as it learns, it is still very difficult to accurately measure epigenetic changes during cellular development. Not enough cells can usually be obtained that are at precisely the same stage for scientists to map specific molecules at specific chromosomal locations."
Allan Spradling PhD, Howard Hughes Medical Institute, Department of Embryology, Carnegie Institution for Science, Baltimore, Maryland .
Lee and Spradling took advantage of the genetic tools made available from the sudy of fruit flies to gain insight into follicle cell epigenetics. Using a variety of tools and techniques, they focused on cells in the fruit fly ovary to identify a gene called Lsd1. They found that the Lsd1 gene helps progenitor cells mature at a normal rate while on their way to becoming follicle cells in the fly ovary.
More importantly, they
found that the amount of Lsd1 protein decreases as the cells mature into ovarian follicles. What's more, the onset of differentiation could be changed by changing the level of Lsd1. It appears that differentiation begins when the Lsd1 protein falls below a critical level.
"The timing of differentiation is very important for normal development. Its' onset determines how many times progenitor cells divide, and even small perturbations in Lsd1 levels changed the number of follicle cells that were ultimately produced, which reduced ovarian function."
Ming-Chia Lee PhD, Howard Hughes Medical Institute, Department of Embryology, Carnegie Institution for Science, Baltimore, Maryland
Previously, it was thought that follicle progenitor cells began to differentiate based on a signal from the next egg to erupt within the ovary. Lee and Spradling found that while this germ cell (or egg) signal is essential, the signal changes based on Lsd1 level.
As is frequently the case in basic biological research, molecules and mechanisms studied are found throughout the animal kingdom, including in humans. The importance of this research is understanding how animal chromosomes change during normal development. It may also help clarify alterations in the epigenetic state taking place in some cancers as a minority of cells in those cancers begin to express high levels of Lsd1 and behave like undifferentiated progenitor cells.
"Studying fruit fly follicle cell differentiation can teach us at a deeper level what Lsd1 is doing in both normal and cancerous progenitors."
Ming-Chia Lee PhD
Abstract
DNA replication remains unfinished in many Drosophila polyploid cells, which harbor disproportionately fewer copies of late-replicating chromosomal regions. By analyzing paired-end high-throughput sequence data from polytene larval salivary gland cells, we define 112 underreplicated (UR) euchromatic regions 60–480 kb in size. To determine the effects of underreplication on genome integrity, we analyzed anomalous read pairs and breakpoint reads throughout the euchromatic genome. Each UR euchromatic region contains many different deletions 10–500 kb in size, while very few deletions are present in fully replicated chromosome regions or UR zones from embryo DNA. Thus, during endocycles, stalled forks within UR regions break and undergo local repair instead of remaining stable and generating nested forks. As a result, each salivary gland cell contains hundreds of unique deletions that account for their copy number reductions. Similar UR regions and deletions were observed in ovarian DNA, suggesting that incomplete replication, fork breakage, and repair occur widely in polytene cells. UR regions are enriched in genes encoding immunoglobulin superfamily proteins and contain many neurally expressed and homeotic genes. We suggest that the extensive somatic DNA instability described here underlies position effect variegation, molds the structure of polytene chromosomes, and should be investigated for possible functions.
This work was funded in part by the Howard Hughes Medical Institute.
The Carnegie Institution for Science is a private, nonprofit organization headquartered in Washington, D.C., with six research departments throughout the U.S. Since its founding in 1902, the Carnegie Institution has been a pioneering force in basic scientific research. Carnegie scientists are leaders in plant biology, developmental biology, astronomy, materials science, global ecology, and Earth and planetary science.
Return to top of page
|
