|
|
|
Home | Pregnancy Timeline | News Alerts |News Archive May 11, 2015
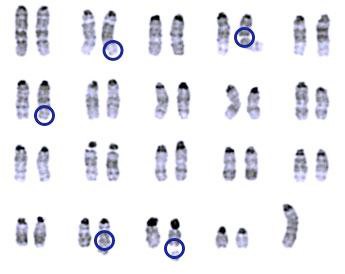
Retrotransposons are ancient remnants of viruses. Their DNA is first transcribed into an
RNA template, then reverse transcribed into DNA — then inserted into a new genomic site.
They jump around more frequently within genomes of cells without the Histone H3.3.
(ABOVE) These cells have abnormalities in their chromosomes (circled) and researchers
speculate H3.3 may help prevent such potentially damaging alterations.
Image Credit: Laura Banaszynski PhD |
|
|
|
|
|
How to stop "jumping genes" in stem cells
Histone H3.3 helps keep the mouse genome stable
by keeping retrotransposons from "jumping" out of place.
Proteins known as histones provide support and structure to DNA, wrapping the DNA molecule around numerous histone "spools" in order to compact DNA to fit within the nucleus of a cell. But for years, scientists puzzled over occasional "extreme" histones which exist for unexplained reasons. Now, research has uncovered a new purpose for one histone variation (or variant) — the prevention of genetic mutations. This histone variant appears to be keeping some "jumping genes" from jumping out of sequence.
The research began at Rockefeller University and was published May 4, 2015 in the journal Nature. Due to the close relationship between histones and DNA, scientists have known for some time that histones frequently are involved in the epigenetic control of genes. One particular histone variation (variant) appears to reduce the chance of harmful changes in stem cells that could affect various types of tissue making up a living creature.
"They say that good things come in small packages. Nowhere is this more true than with histone variants. This study found the variant of one histone — H3.3 — differs only slightly from the standard H3 histone. It helps prevent certain genetic elements, remnants left behind by ancient viral infections, from moving about within the genome."
C. David Allis, study author Joy and Jack Fishman Professor and head of the Laboratory of Chromatin Biology and Epigenetics
Chemical modifications to histones that spool DNA can change how genes are made active (expressed), or inactivated (silenced) simply by unwinding DNA so that other molecular chemicals can attach and "read" that portion of DNA to make a protein — or stay tightly wound and inaccessible. However, a histone called H3.3 varies from its regular counterpart H3 by only a few amino acids, yet is present throughout the animal kingdom. Scientists have suspected for awhile that H3.3 has some specific, yet at the time, unknown job.
Study authors Simon Elsasser and Laura Banaszynski began working on H3.3 in David Allis's lab at Rockefeller University. They isolated H3.3 in the mouse genome of stem cells. It was Elsasser's idea to look for H3.3 in repetitive sequences of genes. However, repeats are normally filtered out in a genome-wide study, so Elsasser developed a new approach to capture the repeats.
And a pattern did emerge. Histone H3.3 appeared in retrotransposons, the leftovers of ancient viral infections. They are trapped in their genome host, but can still copy themselves and jump to new locations within that genome. Sometimes for a very good reason. Retrotransposon derived genes provided the code for proteins necessary to form the placenta in mammals. But when retrotransposons jump, they can sometimes interrupt needed gene codes and cause harmful mutations.
Stem cell chromasomes are more plastic than those found in differentiated cells because stem cells have more capacity to become hundreds of different cell types needed in an organism. But once a cell has picked a tissue identity, un-needed parts of that genome are turned off forever.
Prior to the current study, scientists knew mouse stem cells kept control of retrotransposons by tagging the H3 histone with chemical markers containing three methyl groups.
Early experiments done by Laura Banaszynski suggested H3.3 was needed to correctly place those suppressive "trimethyl" marks. Banaszynski: "By taking away proteins responsible for placing H3.3 into chromatin, or eliminating H3.3 completely, we confirmed that trimethylation depends on H3.3. This study hints at a fascinating question in biology — How do cells balance the potential evolutionary benefit of mobile elements, such as retrotransposons, with the competing need to silence them so as to maintain the genome?" "
But Simon Elsasser ponts out that more experiments need to be done as : "... we have not eliminated the possibility that loss of H3.3 results in genomic instability for other reasons."
Although the types of retrotransposons studied in these experiments are not active in humans, it's likely that human stem cells have their own H3.3 histone to keep other varieties of human jumping genes in place. This research, therefore, has implications beyond epigenetics.
Abstract
Transposable elements comprise roughly 40% of mammalian genomes1. They have an active role in genetic variation, adaptation and evolution through the duplication or deletion of genes or their regulatory elements 2, 3, 4, and transposable elements themselves can act as alternative promoters for nearby genes, resulting in non-canonical regulation of transcription 5, 6. However, transposable element activity can lead to detrimental genome instability 7, and hosts have evolved mechanisms to silence transposable element mobility appropriately 8, 9. Recent studies have demonstrated that a subset of transposable elements, endogenous retroviral elements (ERVs) containing long terminal repeats (LTRs), are silenced through trimethylation of histone H3 on lysine 9 (H3K9me3) by ESET (also known as SETDB1 or KMT1E)10 and a co-repressor complex containing KRAB-associated protein 1 (KAP1; also known as TRIM28)11 in mouse embryonic stem cells. Here we show that the replacement histone variant H3.3 is enriched at class I and class II ERVs, notably those of the early transposon (ETn)/MusD family and intracisternal A-type particles (IAPs). Deposition at a subset of these elements is dependent upon the H3.3 chaperone complex containing α-thalassaemia/mental retardation syndrome X-linked (ATRX)12 and death-domain-associated protein (DAXX)12, 13, 14. We demonstrate that recruitment of DAXX, H3.3 and KAP1 to ERVs is co-dependent and occurs upstream of ESET, linking H3.3 to ERV-associated H3K9me3. Importantly, H3K9me3 is reduced at ERVs upon H3.3 deletion, resulting in derepression and dysregulation of adjacent, endogenous genes, along with increased retrotransposition of IAPs. Our study identifies a unique heterochromatin state marked by the presence of both H3.3 and H3K9me3, and establishes an important role for H3.3 in control of ERV retrotransposition in embryonic stem cells.
Return to top of page
|
