|
|||||||||||||||
|

CLICK ON weeks 0 - 40 and follow along every 2 weeks of fetal development
|
||||||||||||||||||||||||||||
|
Fetal Timeline Maternal Timeline News News Archive Aug 11, 2015
|
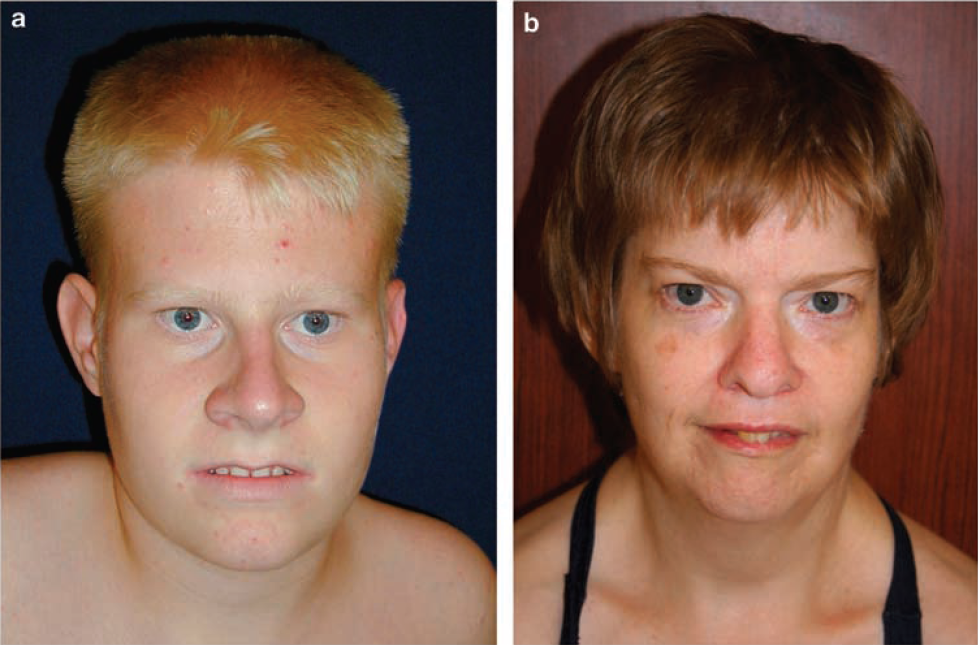
How a single genetic mutation can cause autism Last December it was announced that more than 1,000 gene mutations in people with autism had been identified, but it was left unexplained how these particular mutations increased the risk for autism. Now, researchers at the University of North Carolina (UNC) School of Medicine have found how one disabled molecular switch in one gene can lead to the disorder, while duplication or triplication of the same switch can lead to Angelman Syndrome (AS). AS is a classic example of genomic imprinting. It is caused by deletion or inactivation of genes on the mother's chromosome 15. The father's copy of chromosome 15, which may be normal, is imprinted and therefore silenced. The companion syndrome, Prader-Willi Syndrome, is caused by a similar loss of a father's inherited genes and a mother's genes being imprinted and also silenced. The research is published in the journal Cell
Research was conducted on human cell lines and in mouse models, as Zylka had cells from previous research with an autism-linked UBE3A mutation patient. Sequencing genes from those cell samples - including cells from the child's parents - Zulka found that only the child had hyperactive UBE3A, not the parents. The child's regulatory switch was broken causing UBE3A to be perpetually switched on and ubiquitin not being attached to proteins needing to be destroyed. "When this child's mutation was introduced into an animal model, we saw all these dendritic spines form on the [mouse's] neurons," explained Zylka, a member of the Carolina Institute for Developmental Disabilities. "Having too many dendritic spines has been linked to autism." Their findings thus pointed to hyperactivation of UBE3A as the likely cause of the child's condition. Duplication of the 15q chromosome region - which includes UBE3A and several other genes - is one of the most commonly seen genetic alterations in people with autism and is called "Dup15q Syndrome."
One of the drugs, rolipram, previously had been tested in clinical trials treating depression but was discontinued when a patient suffered sudden death by epileptic seizure. In light of this result, Zylka believes retesting must be done with lower doses of rolipram, or any other drug that may increase PKA activity, if only to provide symptom relief for some Dup15q individuals. "The benefits might outweigh the risks," he feels. Such tests would be done with an animal model of Dup15q first. While the bulk of this project focused on autism, it began by identifying Angelman Syndrome-linked mutations clustered in the same chromosome region where a phosphate group attaches to UBE3A. Zylka's team found that a number of Angelman mutations disrupt the function of UBE3A — and that deleting UBE3A function altogether causes cell chaos. Mutations that could essentially eliminate the UBE3A enzyme in Angelman Syndrome patients, is a discovery which could also help diagnose people with this rare and often misdiagnosed disorder. Abstract Highlights Other co-authors included Ben Philpot, PhD, professor of cell biology and physiology, William Snider, MD, director of the UNC Neuroscience Center, and Klaus Hahn, PhD, the Ronald Thurman Distinguished Professor of Pharmacology. Janet Berrios, a graduate student at UNC, and Jason Newbern, PhD, assistant professor in the School of Life Sciences at Arizona State University, are also co-authors. The National Institutes of Health, the Angelman Syndrome Foundation, The Foundation for Angelman Syndrome Therapeutics, and Autism Speaks funded this work.
|
||||||||||||||||||||||||||||