|
|||||||||||||||
|

CLICK ON weeks 0 - 40 and follow along every 2 weeks of fetal development
|
||||||||||||||||||||||||||||
|
Fetal Timeline Maternal Timeline News News Archive Aug 26, 2015
|
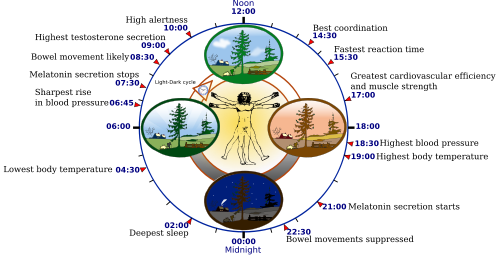
Algorithm identifies gene movement around the clock Genes that turn on in precisely timed patterns are known as oscillatory genes. They are essential to cell division, limb formation, and control our circadian rhythms. However, without us having a time-lapse view of when these genes begin to function, oscillatory genes can go unidentified and delay our understanding of molecular functions impacting development. Now, a study published in Nature Methods describes a new statistical approach, called "Oscope," to help identify oscillating genes. The key to Oscope is examining unsynchronized cells during different cell states - compare results and average when the possibility for oscillation will begin in each. Identifying oscillatory genes using traditional RNA-sequencing requires scientists to choose a known system and identify cells as they synchronize to that system. But this process may mask different types of oscillatory signals developing later, from other signals sychronizing sooner. With Oscope "we study unsynchronized single cell data so none of the oscillatory systems are disturbed," says Ning Leng, computational biologist with the Morgridge Institute at the University of Madison Wisconsin-Madison (UW-Madison) and co-author on the paper. "This enables us to discover unknown oscillatory signals."
Statistical expertise in this project was provided by professor Christina Kendziorski from the Department of Biostatistics and Medical Informatics at UW-Madison. Her group worked in collaboration with the cell biology strengths of stem cell research pioneer — James Thomson — and his group at the Morgridge Institute for Research. The Oscope software package is offered free to scientists at: https://www.biostat.wisc.edu/~kendzior/OSCOPE/.
The goal of the Thomson lab is to establish a system to detect timing-related genes important in development. The most well understood oscillatory genes are those related to our circadian clock. These genes become active or inactive in tune with the sun cycle and tightly regulate cell proliferation and metabolism over a 24-hour period. They affect disease, the most notable being cancer where cell division is hijacked by cells proliferating out of control. The Oscope technique may help scientists identify the connections between gene cycles and disease cycles. Although single-cell RNA sequencing is an extremely powerful tool for getting a snapshot of when genes become active, it is less able to trace a single gene over time. Oscope uses high-resolution snapshots of data to identify different gene groups that are potentially changing over time and possibly capture oscillating genes in that group.
Abstract
|
||||||||||||||||||||||||||||