|
|
Fetal Timeline Maternal Timeline News News Archive Sep 14, 2015
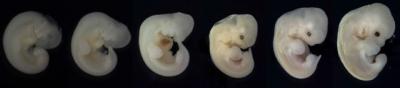
Embryo of the buffy flower bat, showing progressive growth of its' limbs
Image Credit: Courtesy of Karen Sears
|
|
| |
|
|
|
What steers evolution? Genes, then nature, then genes..
Natural selection is a race to reproduce, a competition between individuals affecting evolution within a species. Scientists exploring networks that shape individual form and function, now ask: How do genes determine which individuals get to compete, before evolutionary competition even begins?
University of Illinois researchers Karen Sears and Zoi Rapti, along with collaborators from four other institutions, addressed this question by following gene networks guiding limb development in mammals.
They found during early development, when limbs just begin to bud and extend from the body, gene activity is pretty stable. But when specific details — knees, elbows, wrist or ankle bones, digits, claws, or even thickness of hair and skin — gene activity goes wild. This pattern may reflect adaptation of the later embryo to environmental changes such as temperature and rainfall which influence the growth of plants, and the nutrient levels changing within mom's diet. Nature, rather than remodel basic limb structure, evolves the later fetus through numerous, small tweaks to fit into its new world.
"When we look at the evolutionary record of animals, we find there are some forms that have evolved repeatedly, and some that have never evolved. I want to know the role development has in generating these different patterns."
Karen Sears PhD, Associate Professor, Animal Biology, Carl R. Woese Institute for Genomic Biology, University of Illinois, Urbana-Champaign
Sears is a member of the Carl R. Woese Institute for Genomic Biology (IGB) and Rapti is an associate professor of mathematics. They led a study published in PLOS Genetics looking for gene patterns of expression to explain what happens when a mutation changes the activity of one gene. How much will the whole network and limb be affected?
Many genes code proteins that either influence or specifically regulate other genes. These connections could be compared to the threads and intersections of a spider's web. Some interactions are stronger than others, and some genes have more connections than others. Mathematics can help model these gene networks and assist in predicting the outcomes of network interruptions. Building on published data, the scientists created a computer map of how genes might interact during early and late stages of limb development.
This map allowed them to pluck threads of the gene web and watch what happened. They found in early limb development, the network resisted spreading any changes. If a gene was altered, the network continued to function unaffected. However, later in limb development the network responded immediately. A change in one gene had a widespread impact.
Applying what they had mapped to the actual limb development in four mammals — mice, bats, opossums, and pigs — they found genes in each of the four species differed more in late development than early. Theoretically and through experiments, their findings supported Sears' belief that genes in early limb development have little variation between mammal species.
"If early development is disrupted, limb development will be severely disrupted, and it is very unlikely that the resulting limb will be selectively advantageous to that animal. Later development, which doesn't have as many downstream impacts, might be expected to be more free to vary because consequences of variation would be less dire."
Karen Sears PhD
From an evolutionary perspective, this makes sense.
Abstract
Variation among individuals is a prerequisite of evolution by natural selection. As such, identifying the origins of variation is a fundamental goal of biology. We investigated the link between gene interactions and variation in gene expression among individuals and species using the mammalian limb as a model system. We first built interaction networks for key genes regulating early (outgrowth; E9.5–11) and late (expansion and elongation; E11-13) limb development in mouse. This resulted in an Early (ESN) and Late (LSN) Stage Network. Computational perturbations of these networks suggest that the ESN is more robust. We then quantified levels of the same key genes among mouse individuals and found that they vary less at earlier limb stages and that variation in gene expression is heritable. Finally, we quantified variation in gene expression levels among four mammals with divergent limbs (bat, opossum, mouse and pig) and found that levels vary less among species at earlier limb stages. We also found that variation in gene expression levels among individuals and species are correlated for earlier and later limb development. In conclusion, results are consistent with the robustness of the ESN buffering among-individual variation in gene expression levels early in mammalian limb development, and constraining the evolution of early limb development among mammalian species.
Author Summary
The variation generating mechanisms of development interact with the variation sorting mechanism of natural selection to produce organismal diversity. While the impacts of natural selection on existing variation have received much study, those of development on the generation of this variation remain less understood. This fundamental gap in our knowledge restricts our understanding of the key processes shaping evolution. In this study, we combine mathematical modeling, and population-level and cross-species assays of gene expression to investigate the relationship between the structure of the gene interactions regulating limb development and variation in the expression of limb genes among individuals and species. Results suggest that the way in which genes interact (i.e., development) biases the distribution of variation in gene expression among individuals, and that this in turn biases the distribution of variation among species.
Sears, Rapti, and coauthors were brought together by the Illinois BioMathematics Program, an NSF-funded project that promotes research collaboration among biology and mathematics undergraduates and faculty members. The BioMath Program has led to a variety of innovative research efforts. Sears is a member of the Regenerative Biology & Tissue Engineering research theme and an affiliate of the Gene Networks in Neural & Developmental Plasticity research theme at the IGB.
Return to top of page
|



