|
|||||||||||||||
|

CLICK ON weeks 0 - 40 and follow along every 2 weeks of fetal development
|
||||||||||||||||||||||||||||
|
Messenger RNA may fine tune protein production Long thought of as a simple link between DNA and protein, new research at The Rockefeller University suggests messenger RNA is up to something unexpected. By uncovering widespread differences in the expression of parts making up the mRNA molecule — something assumed not to happen — scientists think they have found intriguing patterns suggesting functions for regions within mRNA molecules competely unexpected. "The lopsided ratios we found in two parts of mRNA — one that carries the code for a protein and one that doesn't — do not appear to be random," says senior author Mary Hynes, a research associate professor. "We suspect some of these skewed ratios may be regulating protein production, particularly during embryonic development, but also in adults." Reported December 16 in Neuron, the research results focus on regions within messenger RNA that imparted different cell responses.
The 5' untranslated region (5′ UTR) is also known as a Leader Sequence or Leader RNA. It is the region of mRNA directly upstream giving the signal for the beginning of transcription. It specifies the first amino acid in protein synthesis and so is important for regulating transcription. However, using a technique developed at Rockefeller University called translating ribosome affinity purification or TRAP, the Hynes' team isolated purified mRNA from dopamine neurons in mouse embryos. TRAP is a new technique (2014) that shows the entire cell population of mRNA, and reduces it's complexity. Its a view that helps observers understand cell function on the molecular level.
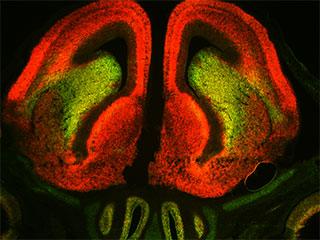
An earlier study noted similar disparities, but did not explore the biological implications and that study's data did not get widespread attention. Hynes and her colleagues took the next step, asking: Why would a cell make abundant levels of 3'UTR sequence with no 5'UTR coding sequence, since protein cannot be made without the coding sequence? To verify their findings and understand if this was restricted to dopamine neurons, development, or just the nervous system, the scientists used green probes to mark 5'UTR coding sequences and red to mark the 3'UTRs of 19 genes in embryonic and adult tissue. "Based on prior understanding, it was expected that every cell in the tissue should show up as either yellow, when both are expressed, or black, when neither are," Hynes says. "But to our surprise, when we examined Sox11 mRNA in the brain we found many neurons that were red, or expressing mostly UTR, as well as many that were green, or expressing mostly coding sequences." They went on to show this was true for every gene examined and that different levels of expression of UTR and coding sequences occurs throughout the embryo, the adult, and outside of the nervous system.
However, it's not clear how this might happen, Hynes adds. Next, they focused back on the developing dopamine neurons and the nine thousand genes they found active in them.
Adds Hynes: "During development a neuron may need to express a certain gene, but only a particular amount. Either too much or too little might be harmful and lead to irreversible changes. So, we think this could be a mechanism for finely titrating [adjusting the balance] of proteins levels in an active gene."
Hynes: "Going forward, I think that when an RNA sequencing experiment suggests that a gene is highly expressed, researchers should take a closer look at the relative levels of these two other components to get a more accurate picture of what is being expressed." |
Dec 25, 2015 Fetal Timeline Maternal Timeline News News Archive
|
||||||||||||||||||||||||||||