|
|
Id tags define developing neural circuits
The human brain is composed of circuits made up of neurons, cells specialized to transmit information via electrochemical signals. Like the circuits in a computer, these neuronal circuits must connect in very unique ways in order to function. But with billions in a single human brain, how does a neuron make the right connection with the right cell?
Biologists have long searched for some kind of cell level "identification tag" that labels which cell forms what connections. The laboratory of Kai Zinn PhD professor of biology at the California Institute of Technology (Caltech) in Pasadena, California, has now found molecular tags in the fruit fly - Drosophila. The molecular id tags discovered were in two different molecular subfamilies - Dpr and DIP.
DNA is a large biomolecule, as is any molecule in a living organism, essential to all known forms of life. It functions to encode, transmit and express genetic information. Zinn's group has now defined how a network of interacting cell surface proteins (a 21-member Immunoglobulin superfamily or IgSF) the Dprs, binds to another nine-member subfamily, the DIPs.
Immunoglobulin superfamily CAMs (IgSF CAMs) are types of Cell Adhesion Molecules or CAMs, and are part of a large group of cell surface proteins that recognize and bind cells together. Molecules are categorized as members of this superfamily based on the structural features they share with immunoglobulins (antibodies).
Dpr and DIP selectively bind together and that bond plays an important role in directing the development of neuromuscular and visual systems in a growing Drosophila. The research is published in the December 17 issue of the journal Cell1.
In 2013, a collaboration between Christopher Garcia's structural biology group at Stanford and the Zinn group at Caltech mapped interactions between all 200 Drosophila cell surface proteins. By observing their interactions in a test tube, researchers determined which proteins bound together. The group then developed a complex model of interacting proteins — calling it the interactome.
Research revealed a 21-member subfamily — the Dprs — an "immunoglobulin superfamily" which selectively binds to a 9-member subfamily of DIPs.
"Certain members of the Dprs and the DIPs match up and bind together— like a lock and key — in a test tube," says Zinn. "We wanted to know if they would bind in vivo, in the Drosophila brain, and if that bond then determined where synapses formed."
A synapse is a junction where the wire-like axon of one neuron meets the branched dendrites of another. Chemical signals — or neurotransmissions — pass between neurons by crossing these synapse junctions.
"We wanted to know if these interacting proteins on the surface of neuronal cells affect the way that cells interact. We showed that neural cells that expressed matching proteins often formed synapses with each other. We theorize that interaction between these molecules drives the formation of synapses."
Robert Carrillo, postdoctoral scholar, the Zinn group, and co-first author on the new paper.
The Zinn group used the well-studied Drosophila visual system to determine the effects of these proteins on its' development. Neurons in the fly's eye send axons into layered structures in the visual part of the brain, known as the optic lobe. One of these structures, the medulla, is divided into ten layers, and each optic lobe neuron forms synapses within a specific subset layer.
By removing certain DIP and Dpr proteins in the fly pupa, the researchers caused axons to "overshoot" their target layers. They also observed developmental defects in a fly's neuromuscular system when the same proteins were removed.
Another paper in the same issue of Cell2, from Larry Zipursky's group at UCLA, also found that expression of Dprs and DIPs correlates with the patterns of synaptic connectivity in the brain.
This finding helps to validate a theory proposed in the 1950s by the late Caltech professor and Nobel Laureate Roger Sperry. Experimenting mostly with fish and frog brains, Sperry discovered that he could manipulate or cut axons between neurons, and the cells would still re-form the right connections.
"Sperry hypothesized that individual neurons must carry some kind of identification tags, whose recognition is used to create synaptic circuits in the brain. Our group has shown that the Drosophila Dpr and DIP proteins fit the definition of Sperry's proposed cellular identification tags."
Kaushiki Menon, senior postdoctoral scholar in the Zinn group and a co-first author on the paper.
Such tinkering with the brain's circuitry is possible because flies, unlike humans, have brains that are predominantly "hard-wired." "In mammals, the brain has a basic initial scaffold laid down by genetics, and then over time there is a lot of complicated experience-dependent rearrangement." Zinn: "Essentially, the human brain can rewire itself through experience. Fly brains can't do that."
While their findings are not immediately generalizable to mammals, Zinn and his group hope that they can provide a starting point to probe the structure of the human brain. Zinn: "We hope that there might be protein networks that function similarly in humans, and these could be relevant to an understanding of how the scaffold of the human brain that exists at birth is assembled through genetics."
Cell 1: Control of Synaptic Connectivity by a Network of Drosophila IgSF Cell Surface Proteins
Abstract Highlights
•Twenty-one Dpr and nine DIP surface proteins form a complex interaction network in Drosophila
•Dprs and DIPs interact via a hydrophobic interface between their first Ig domains
•Each Dpr and DIP is expressed by a unique small subset of larval and pupal neurons
•Loss of Dpr-DIP interactions affects synaptic connectivity in the brain
Summary
We have defined a network of interacting Drosophila cell surface proteins in which a 21-member IgSF subfamily, the Dprs, binds to a nine-member subfamily, the DIPs. The structural basis of the Dpr-DIP interaction code appears to be dictated by shape complementarity within the Dpr-DIP binding interface. Each of the six dpr and DIP genes examined here is expressed by a unique subset of larval and pupal neurons. In the neuromuscular system, interactions between Dpr11 and DIP-γ affect presynaptic terminal development, trophic factor responses, and neurotransmission. In the visual system, dpr11 is selectively expressed by R7 photoreceptors that use Rh4 opsin (yR7s). Their primary synaptic targets, Dm8 amacrine neurons, express DIP-γ. In dpr11 or DIP-γ mutants, yR7 terminals extend beyond their normal termination zones in layer M6 of the medulla. DIP-γ is also required for Dm8 survival or differentiation. Our findings suggest that Dpr-DIP interactions are important determinants of synaptic connectivity.
Reference: McMurtrey RJ. Analytic Models of Oxygen and Nutrient Diffusion, Metabolism Dynamics, and Architecture Optimization in Three-Dimensional Tissue Constructs with Applications and Insights in Cerebral Organoids. Tissue Engineering. doi: 10.1089/ten.TEC.2015.0375
In addition to Carrillo and Menon, structural biologist Engin Ozkan at the University of Chicago is a co-first author.
The paper is titled "Control of synaptic connectivity by a network of Drosophila IgSF cell surface proteins."
Work was funded by the National Institutes of Health and the Howard Hughes Medical Institute.
Cell2: Induction in the developing compound eye of Drosophila: Multiple mechanisms restrict R7 induction to a single retinal precursor cell
Abstract
The development of the Drosophila R7 photoreceptor cell is determined by a specific inductive interaction between the R8 photoreceptor cell and a single neighboring precursor cell. This process is mediated by bride of sevenless (boss), a cell-surface bound ligand, and the sevenless (sev) tyrosine kinase receptor. The boss ligand is expressed specifically on the surface of the R8 cell, whereas the sev receptor is expressed on 5 cells contacting the developing R8 cell and other cells not in contact with R8. By altering the spatial and temporal expression of boss, we demonstrate that sev-expressing cells that do not contact R8 can assume an R7 cell fate. By contrast, the sev-expressing precursor cells to the R1–R6 photoreceptor cells that do contact R8 are nonresponsive to the inductive cue. Using the rough and Nspl mutations, we demonstrate that an early commitment to an R1–R6 cell fate blocks the pathway of sev activation in these cells.
Return to top of page
|
|
|
Jan 5, 2016 Fetal Timeline Maternal Timeline News News Archive
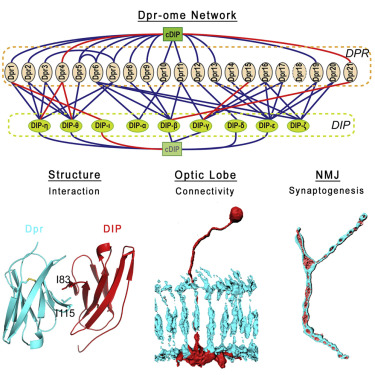
Twenty-one Dpr and nine DIP proteins form a complex network between
Drosophila neurons. The loss of Dpr-DIP interactions affects
synaptic connectivity in the fly brain.
Image Credit:
California Institute of Technology, Pasadena, CA, USA
|
|
|
|



