|
|
Trimmer snips jumping genes
A Pac-Man-like enzyme called "Trimmer," protects sperm and eggs from genetic rewriting by our "jumping genes."
Transposable elements (TEs), also called "jumping genes," are DNA sequences that move from one location on the genome to another. They were first identified more than 50 years ago by Barbara McClintock of Cold Spring Harbor Laboratory in New York. Initially skeptical, biologists over the next several decades found TEs in almost all organisms (prokaryotes and eukaryotes) and in large numbers. TEs make up approximately 50% of our human genome and up to 90% of the maize (corn) genome.
Much of what a transposon does depends on where it lands on a gene. Landing inside a gene can result in a mutation, as in hemophilia. Similarly, in the APC genes in colon cancer cells but not in the APC genes in healthy cells in the same individuals. This confirms that some transposons might cause diseases to develop depending on their placement on the gene. Some silenced TEs are inactive because they have mutations that affect their ability to move from one chromosome location to another. Other silenced TEs are perfectly intact and capable of moving but kept inactive by epigenetic defenses such as DNA methylation, chromatin remodeling, and miRNAs. So, organisms need to keep jumping genes under control. Particularly in germ cells (sex cells) that give rise to sperm and eggs.
"Jumping genes," or transposons, are small segments of DNA that move around within our genome. They can disrupt host genes and have been implicated in cancer and other diseases.
However, the ability of transposons to increase genetic diversity, together with the ability of the genome to inhibit most "jumping gene" activity, results in a balance that makes them an important part of species adaptability in evolution.
Because transposon movement can be destructive, most transposon sequences in the human genome are silent, allowing our genome to remain relatively stable despite their prevalence. Gene regulation in all organisms that carry these sequences, however, must ensure the integrity of future generations from adaptations that may be needed or benign in the current organism, but unneccessary or toxic in the next. Turning off jumping genes in germ cells is done by a class of PIWI-interacting RNAs or small RNAs called piRNAs.
Typically only 24 to 30 nucleotides long, piRNAs stop jumping genes from being turned on — known as expression. But, piRNAs do not become effectve themselves without also being trimmed to a final, "mature" length by pre-piRNAs. However, what pre-trimming enzyme was cutting piRNAs had been unidentified.
Now, research Associate Natsuko Izumi and Professor Yukihide Tomari, University of Tokyo Institute for Molecular and Cellular Biosciences, and colleagues at the Computational Medicine Center, Sidney Kimmel Medical College, Thomas Jefferson University, Philadelphia, Pensylvania, USA, have successfully identified a ribonuclease in silkworm ovary cells as that jumping gene trimming protein. They call it "Trimmer."
Results of this research can be read in Cell.
Their data shows that Trimmer requires Papi, a PIWI-associated protein, to trim off the end of pre-piRNAs. Trimming pre-piRNAs probably takes place on the surface of mitochondria and is critical in turning off "jumping genes".
"We discovered the trimming activity in the pellet of silkworm cells in 2011. Although we knew where the enzyme existed, it was extremely difficult to identify as it is insoluble. In fact, we almost gave up many times.
"A breakthrough came when we noticed trimming activity is enriched in cell mitochondria. We then patiently attempted to solubilize mitochondria trimming activity. It took another three years to identify "Trimmer".
The second breakthrough came when we realized that "Trimmer" partners with Papi."
Yukihide Tomari PhD, Professor, Institute of Molecular and Cellular Biosciences, The University of Tokyo, Bunkyo-ku, Tokyo, Japan
Abstract Highlights
•The exonuclease PNLDC1 is the pre-piRNA 3′ Trimmer in silkworms
•PNLDC1 binds the Tudor protein Papi/Tdrkh on the mitochondrial surface
•Papi/Tdrkh binds PIWI proteins, recruiting PIWI-bound pre-piRNAs to PNLDC1
•Untrimmed pre-piRNAs are impaired for target cleavage and prone to degradation
Summary
PIWI-interacting RNAs (piRNAs) play a crucial role in transposon silencing in animal germ cells. In piRNA biogenesis, single-stranded piRNA intermediates are loaded into PIWI-clade proteins and cleaved by Zucchini/MitoPLD, yielding precursor piRNAs (pre-piRNAs). Pre-piRNAs that are longer than the mature piRNA length are then trimmed at their 3′ ends. Although recent studies implicated the Tudor domain protein Papi/Tdrkh in pre-piRNA trimming, the identity of Trimmer and its relationship with Papi/Tdrkh remain unknown. Here, we identified PNLDC1, an uncharacterized 3′-5′ exonuclease, as Trimmer in silkworms. Trimmer is enriched in the mitochondrial fraction and binds to Papi/Tdrkh. Depletion of Trimmer and Papi/Tdrkh additively inhibits trimming, causing accumulation of ∼35–40-nt pre-piRNAs that are impaired for target cleavage and prone to degradation. Our results highlight the cooperative action of Trimmer and Papi/Tdrkh in piRNA maturation.
About the University of Tokyo
The University of Tokyo is Japan's leading university and one of the world's top research universities. The vast research output of some 6,000 researchers is published in the world's top journals across the arts and sciences. Our vibrant student body of around 15,000 undergraduate and 15,000 graduate students includes over 2,000 international students. Find out more at http://www.u-tokyo.ac.jp/en/ or follow us on Twitter at @UTokyo_News_en.
Funding
Grant-in-Aids for Scientific Research on Innovative Areas "Functional machinery for non-coding RNAs" and "non-coding RNA neo-taxonomy"
Reference article: Scitable by Nature Education: "Transposons: The Jumping Genes"
Return to top of page
|
|
|
Mar 3, 2016 Fetal Timeline Maternal Timeline News News Archive
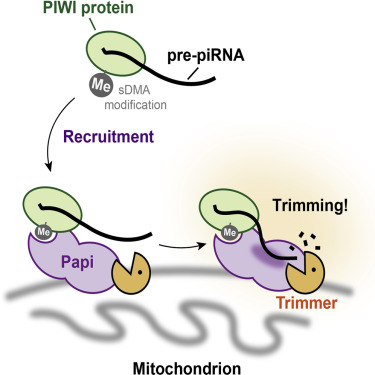
Image Credit: Tomari lab, Institute of Molecular and Cellular Biosciences,
University of Tokyo, Bunkyo-ku, Tokyo, Japan; and Cell
magazine.
|
|
|
|



